- Although L. monocytogenes is a single species of Listeria, small changes in DNA mean there are a range of slightly different L. monocytogenes, which we call strains.
- Because of these small differences, strains of L. monocytogenes can be identified via microbiological typing. This may be particularly useful when L. monocytogenes is isolated at various times or places in a manufacturing environment, to check whether they are the same strain, which would indicate plant persistence.
- Typing may be expensive, and the number of labs which can offer typing is more limited than standard L. monocytogenes detection.
- Methods of typing are explored further below, which include; antibodies, whole genome sequencing, PCR and PFGE.
Typing
In summary
Microbiological typing of strains
The term Listeria monocytogenes is used to refer to a range of closely-related bacteria. Although the bacteria are similar in terms of their DNA and the genes that their DNA contains, an important point is that all L. monocytogenes strains are not identical. Bacterial typing is the process of distinguishing between different, closely-related strains. Typing is important for processors that have isolated L. monocytogenes from final product or their plant environment on different occasions, and that are concerned they may have L. monocytogenes plant resident strains. Sophisticated typing allows two or more L. monocytogenes isolates to be compared to determine if they are the same or different.
It should be made clear that comparative typing of L. monocytogenes is not a routine laboratory test and it is likely to be expensive. It is likely that typing will be beyond the capabilities of most commercial contract laboratories and thus will need to be undertaken by a specialist lab e.g. a university research lab or a government or NHS reference laboratory.
There are a number of different ways to undertake bacterial typing, which include:
- The use of antibodies for serovar determination. Although fairly inexpensive, the method is not very discriminatory. However, some contract labs may offer this as a service. The method is good enough for indication, but not a definitive assessment of the presence of persistent strains.
- The most sophisticated, discriminatory method is whole genome sequencing (WGS). WGS obtains the DNA sequence for a whole bacterium (or sometimes just the important parts of the whole genome) and has recently become much cheaper compared with historical pricings of several thousands of pounds per sample. A whole bacterial genome can be shotgun sequenced for a few hundred pounds. As the cost of WGS has fallen, it has become more popular (Madden et al. 2018) in the investigation of food safety issues (Pietzka et al., 2019). More recently, the highly discriminatory technology has been used to follow mutations in the DNA of L. monocytogenes as it evolves and becomes better adapted to its niche in fish processing environments (Harrand et al. 2020).
- There are alternative genetics-based methods such as polymerase chain reaction (also called PCR, which is inexpensive) or pulsed field gel electrophoresis (PFGE, which was considered for three decades to be the most cost effective and discriminatory typing protocol).
- PFGE is still the preferred typing method by many businesses and research laboratories because if it is done properly, it can allow comparisons to historically typed strains contained within the Pulsenet database. Pulsenet is a global database of food and clinical isolates that is more than 20 years old. The Pulsenet protocols are used by the US-FDA, who routinely test foods such as Scottish cold smoked salmon imported into the USA and type L. monocytogenes isolates. The database is a history of PFGE typed strains that have caused human illness and contaminated foods around the world all of which can be directly viewed and compared if the laboratory test is undertaken to comply with a standardised protocol.
An overview of how PFGE works would make clear that all bacterial cells contain a large molecule called DNA. Genes on the DNA are blueprints for all the activities the cell undertakes (e.g. breaking down sugar to make energy). In essence, PFGE takes the entire DNA inside a cell and cuts it using molecular scissors, which reproducibly cut only at specific places on the DNA. Normally DNA exists as a single, highly twisted, loop in bacterial cells. However, after the DNA is cut it becomes multiple strands that are smaller. These small strands can be separated on the basis of their size (because DNA is negatively charged and moves towards a positively charged electrode). An example of a typical PFGE result is shown as Figure 1.
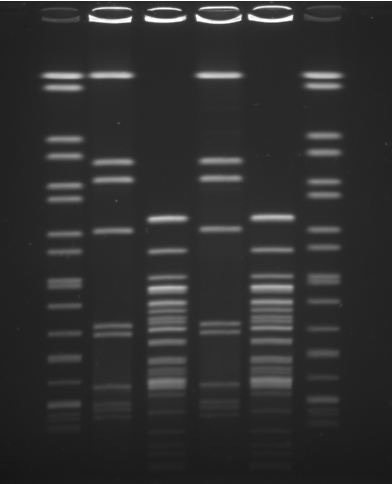
The lane bandings are: Lanes A and F – molecular weight standards of known size, Lane B – L. monocytogenes isolate one cut with ‘scissors’ called AscI, Lane C – L. monocytogenes isolate one cut with ‘scissors’ called ApaI, Lane D – L. monocytogenes isolate two cut with ‘scissors’ called AscI, Lane E – L. monocytogenes isolate two cut with ‘scissors’ called ApaI. The two compared strains are indistinguishable.
An important consideration when typing is to remember that bacteria can evolve to become better suited to their environments over time. Although all organisms (including humans) do that, a bacterial generation can be less than an hour. Thus, there can be small changes to the DNA fingerprints over time as bacteria evolve to become better suited to the niches [glossary entry] they inhabit in processing plants. A specialist typing lab will be able to use mathematical methods to determine whether a single strain isolated from two places has evolved from a common ancestor to generate different fingerprints over time. For that reason, if PFGE typing is undertaken, it is a good practice to use two different pairs of molecular scissors, which cut the DNA in different places, as is shown as Figure 1.
Typing can be complex, and the above explanation is only a short outline of all the issues and how the process can work. It is recommended that processors with suspected resident L. monocytogenes strain(s) contact their Local Authority for information on a specialist typing laboratory for additional technical advice and assistance.
References:
- Harrand, A.S., Jagadeesan, B., Baert, L., Wiedmann, M. and Orsi, R.H. (2020) Evolution of Listeria monocytogenes in a Food Processing Plant Involves Limited Single-Nucleotide Substitutions but Considerable Diversification by Gain and Loss of Prophages. Applied and Environmental Microbiology. 86.
- Madden, R. H., Hutchison M. L., Jordan, K., Pennone, V., Gundogdu, O. and Corcionivoschi, N. 2018. Prevalence and persistence of Listeria monocytogenes in the premises and products of small food business operators in Northern Ireland. Food Control. 87:70-78.
- Pietzka, A., Allerberger, F., Murer, A., Lennkh, A., Stoger, A., Rosel, A.C., Huhulescu, S., Maritschnik, S., Springer, B., Lepuschitz, S., Ruppitsch, W. and Schmid, D. (2019) Whole Genome Sequencing Based Surveillance of L. monocytogenes for Early Detection and Investigations of Listeriosis Outbreaks. Frontiers in Public Health 7, 8.
